IMProve: A Data-Driven Approach to Predicting Integral Membrane Protein Expression
Integral membrane proteins (IMPs) are of both fundamental scientific and medical importance, because they govern the flow of nutrients and information across cell membranes and because they comprise the largest class of therapeutic drug targets. However, the structural and biophysical characterization of IMPs lags far behind that of other protein classes, due to the inability to reliably produce significant quantities of IMPs for experimental studies. We are developing methods to predict and improve IMP expression to address this problem by removing fundamental gaps in knowledge and technology via a combination of computational and experimental methods.
Publications
Decoding sequence-level information to predict membrane protein expression
Shyam M Saladi, Nauman Javed, Axel Müller, William M Clemons Jr.Journal of Biological Chemistry. 2018 Jan 29; 293(13):4913-4927
doi: 10.1074/jbc.RA117.001052
Abstract
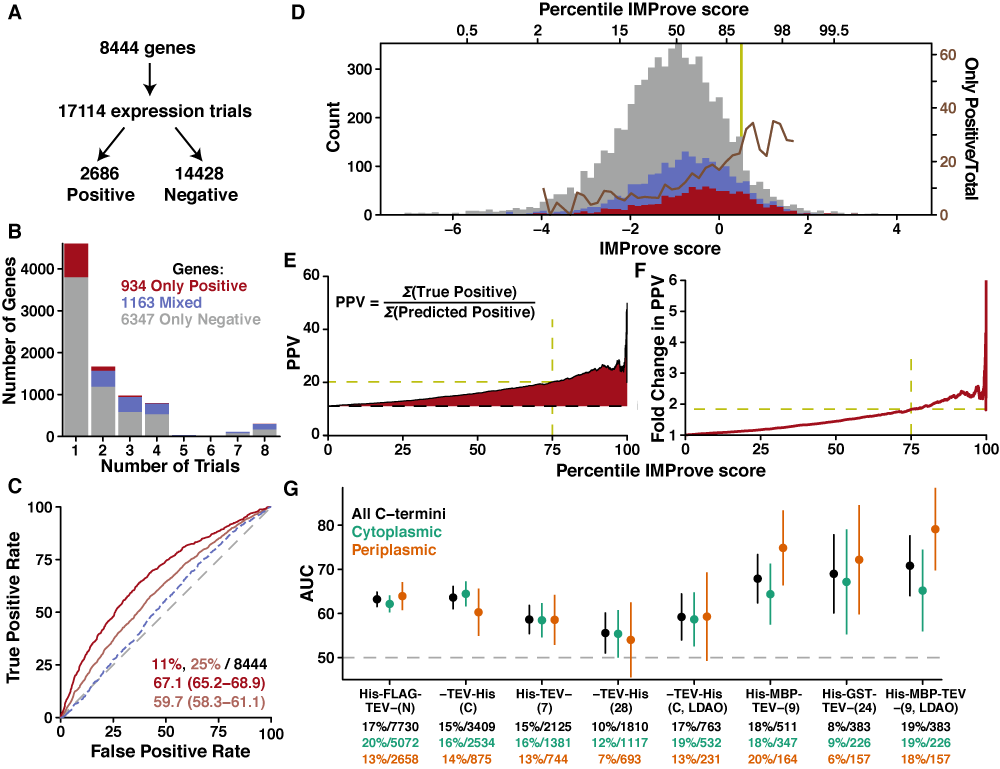
The heterologous expression of integral membrane proteins (IMPs) remains a major bottleneck in the characterization of this important protein class. IMP expression levels are currently unpredictable, which renders the pursuit of IMPs for structural and biophysical characterization challenging and inefficient. Experimental evidence demonstrates that changes within the nucleotide or amino acid sequence for a given IMP can dramatically affect expression levels, yet these observations have not resulted in generalizable approaches to improve expression levels. Here, we develop a data-driven statistical predictor named IMProve that, using only sequence information, increases the likelihood of selecting an IMP that expresses in Escherichia coli. The IMProve model, trained on experimental data, combines a set of sequence-derived features resulting in an IMProve score, where higher values have a higher probability of success. The model is rigorously validated against a variety of independent data sets that contain a wide range of experimental outcomes from various IMP expression trials. The results demonstrate that use of the model can more than double the number of successfully expressed targets at any experimental scale. IMProve can immediately be used to identify favorable targets for characterization. Most notably, IMProve demonstrates for the first time that IMP expression levels can be predicted directly from sequence.
Visit Article Try Model View CodeA Link between Integral Membrane Protein Expression and Simulated Integration Efficiency
Stephen S. Marshall, Michiel J.M. Niesen, Axel Mueller, Katrin Tiemann, Shyam M. Saladi, Rachel P. Galimidi, Bin Zhang, William M. Clemons, Jr., Thomas F. Miller IIICell Reports. 2016 Aug 23; 16(8):2169-2177
doi: 10.1016/j.celrep.2016.07.042
Abstract
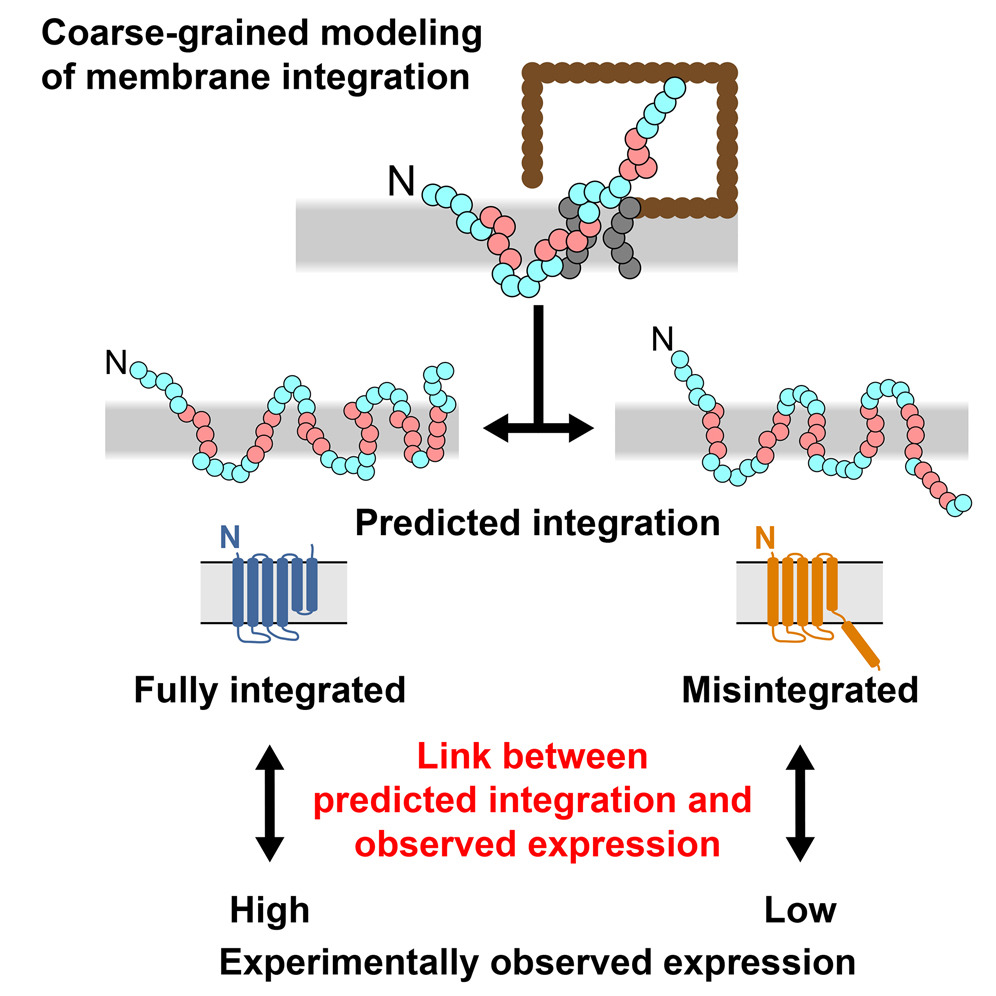
Integral membrane proteins (IMPs) control the flow of information and nutrients across cell membranes, yet IMP mechanistic studies are hindered by difficulties in expression. We investigate this issue by addressing the connection between IMP sequence and observed expression levels. For homologs of the IMP TatC, observed expression levels vary widely and are affected by small changes in protein sequence. The effect of sequence changes on experimentally observed expression levels strongly correlates with the simulated integration efficiency obtained from coarse-grained modeling, which is directly confirmed using an in vivo assay. Furthermore, mutations that improve the simulated integration efficiency likewise increase the experimentally observed expression levels. Demonstration of these trends in both Escherichia coli and Mycobacterium smegmatis suggests that the results are general to other expression systems. This work suggests that IMP integration is a determinant for successful expression, raising the possibility of controlling IMP expression via rational design.
Visit ArticleTeam
Contact us!
We are actively developing this work and are working to make methods accessible to the broader community to accelerate progress of membrane protein biochemical and biophysical studies. Please contact us to discuss early access to new tools, or sign up for an account to be kept abreast of improvements and new releases.





